How to Define a System subject to Mutual Exchange:

Choose Simulate > New.
Define the whole molecule as a a single spin system.
In the absence of exchange, it's possible to
factorize the problem into spin systems, corresponding to the fragments of the molecule.
This is no more possible when iNMR has to simulate the exchange:
in this case each page (still called “system”) describes an exchanging site.
All the “systems” must have an identical number of spins;
apart lucky exceptions, this restriction means that a single document can simulate a single exchange only.
Let's consider the familiar DMF.
Both methyl groups must be defined into the same page,
despite the fact that they don't belong to the same spin system, chemically speaking.
You should define a system of 2 protons with no coupling.
Defining 6 nuclei would require much more computing time and the calculations would also
be less precise (no computers is perfect when dealing with real numbers).
Though not a necessity, at this stage you can close the dialog to verify that the simulation, before introducing the exchange, is correct. Then reopen the dialog with Simulate > Define Systems.
Click the button duplicate. This creates and shows a new system identical to the first one.
If the exchange moves the nucleus A in the position of B (and vice versa) select A and B from the menus at the bottom.
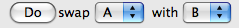
Click the button Do. All the values (shifts and couplings) into the the two rows A and B will be exchanged.
If other nuclei exchange, swap them too. Do this in the second system only (the copy).
Close the dialog and choose Simulate > Dynamic: a new parameter (k12) appears at the bottom of the list, at the left of the plot. It's the rate of exchange (in sec-1).
Select k12 and use the little arrows above the list to increase it. If this is too slow, you can edit the value directly, or increase the value of step.
As long as the little arrows are clicked, the value of step is continuously added or subtracted from the rate of exchange and the plot updated in real time.